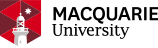Our projects
Our research focuses on platform technology that utilizes the rationally designed multifunctional nanomaterials and sensor platform for biomarkers sensing and cancer molecular subtyping, with the aim to enhance the point-of-care diagnostics and personalized Nanomedicine.
Multifunctional Nanomaterials: Design, Synthesis and Characterization
Multifunctional nanomaterial is of significant interest due to their combination of relevant, desirable features of different materials to form a new material having a broad spectrum of desired properties. By rational design and tailoring the structure of nanomaterials in terms of the size, shape and composition, the properties of the designed nanomaterials could be controlled, in particular for optical property and chemical stability. One typical example is to use the plasmonic nanomaterials (gold or silver) as electrochemical and surface-enhanced Raman spectroscopy (SERS) substrate for signal amplification. Within this research theme, we aim to synthesize multifunctional nanomaterials by using wet-chemistry and further characterize them by electron microscope, e.g. transmission electron microscopy (TEM), UV-vis spectroscopy, electrochemistry and Raman spectroscopy.
Publications:
- Journal of Physical Chemistry C, 128(32), 13518-13528 (2024)
-Journal of the American Chemical Society, 146(9), 5916-5926 (2024)
-Molecular Systems Design and Engineering, 8(2), 251-260 (2023)
-Materials Today Chemistry, 33, 1-10. Article 101698 (2023)
-Nanoscale, 15(5), 2087-2095 (2023)
-Langmuir, 39(44), 15828-15836 (2023)
Nano-Liquid Biopsy for Cancer Diagnostics and Personalised Medicine
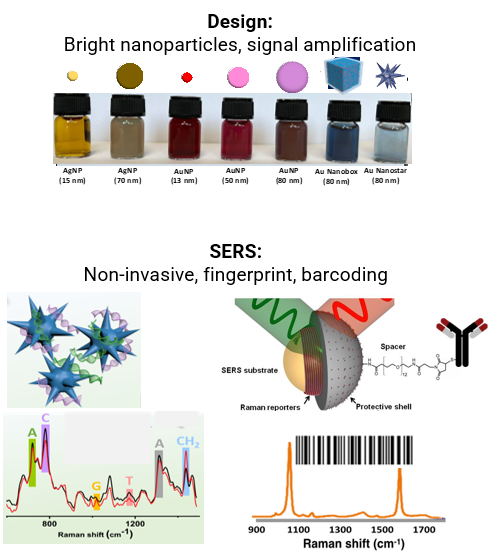
Cancer is a heterogeneous disease which manifests as different molecular subtypes due to the complex nature of tumor initiation, progression and metastasis. Non-invasive analysis of cancer biomarkers such as proteins, genes and cells from body fluids is in high demand and important for cancer diagnostics and treatment as it is the key to achieve the original and intact information from the cells. Thus non-invasive analysis has the benefit of rapid, low-cost and little inconvenience to the patient. Recent advance of nanotechnology has facilitated the application of plasmoinc nanomaterials and surface-enhanced Raman spectroscopy (SERS) in biology and life science due to its unique advantages in the capability of providing ‘fingerprint’ of molecule, multiplexing capability, photo stability, and high sensitivity.
By rational design of a suite of multifunctional nanoparticles (NPs) and taking advantages of the sensor platform in fast sample preparation, we target on simultaneously and selectively detecting a broad panel of liquid biopsy cancer biomarkers (e.g. circulating tumor cells-CTC, circulating tumor DNA-ctDNA and extracellular vesicles-exosomes) with the aim to enable the early cancer diagnosis and personal nanomedicine.
Publications:
-Advanced NanoBiomed Research , 5(6), 1-21. Article 2500008 (2025)
-Nanoscale, 17(7), 3635-3655 (2025)
-The Analyst, 150(10), 2108-2117 (2025)
-Analytical Methods, 14(23), 2255-2265 (2022)
Collaborations: A/Prof Simon Tsao (University of Melbourne and Austin Hospital), A/ProfDhanusha Sabanathan (Macquarie University, Health Medical Oncology and Clinical Trial).
A new molecular tool for direct reading of chemical modifications on DNA
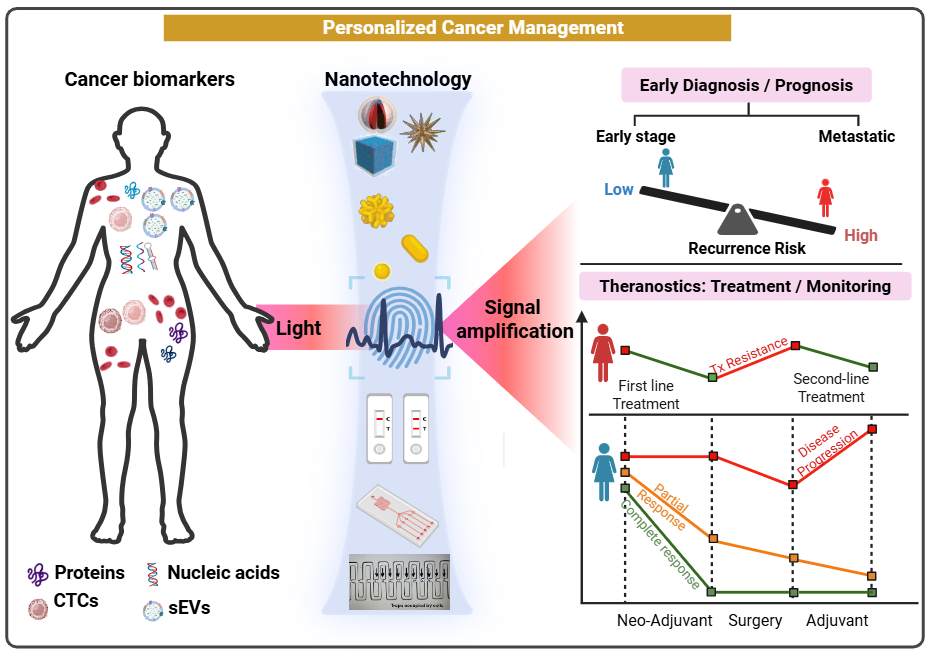
Chemical modifications of DNA and RNA nucleotides (e.g. DNA methylation) affect cellular processes and control genes or genetic programs by turning them “on” and “off”. Among them, the most commonly modified base on DNA is cytosine, which can have additional functional groups added. Typically, the enzymatic addition of a methyl group at the fifth position of the cytosine ring to make 5-methylcytosine (5mC) is the most abundant modified base of the genome. Since the methyl group can also be removed from 5mC via demethylation pathways. This base modification represents one of the most important cellular regulatory mechanisms for controlling gene expression, protein output, and genomic stability. Furthermore, 5mC, and its derivatives 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) have all recently been discovered to play key roles in the regulation of the dynamic balance between DNA methylation and demethylation, mediated by ten-eleven translocation (TET) and thymine DNA glycosylase (TDG) proteins. Due to the different amount of chemical information encoded in different positions/frequencies of methylation on genomic DNAs (referred as DNA methylation landscape), sensitive and accurate analysis of DNA methylation/demethylation can be described as a “Holy Grail” in cell biology, disease states and clinic diagnosis. This project aims to develop a new molecular tool to directly and dynamically read chemical modifications on genomic DNA (epigenetics) by utilizing advanced nanomaterials with the unique features of Raman spectroscopy. Epigenetics affect cellular processes and control genetic programs by turning them “on” and “off" but there is currently no method of direct reading. This project will design a new technology that avoids complicated procedures/chemistry for DNA epigenetic analysis to provide a specific molecular fingerprint of DNA epigenetics.
Publications:
-Chemical Communications, 51(54), 10953-10956 (2015)
-Chemical Communications, 52(17), 3560-3563 (2016)
-Nanotheranostics, 4(4), 224-232 (2020)
-Molecules, 30(2), 1-12. Article 403 (2025)
-The Analyst, 149(10), 2898-2904 (2024)
Collaboration with Prof Alison Rodger (The Australian National University); Prof Joseph Irudayarau (UIUC, USA) and Dr Simon Spencer (The University of Warwick).
Single cell glycomis: mapping the cell surface glycans by using SERS nanotags
Cell surface glycans (sugars, carbohydrates) cover every cell surface and are known to be integral to many cell functions, cell-to-cell interactions and play a role in critical diseases, e.g. cancer and microbial infection. Thus, detecting, imaging and spatial mapping of the density and distribution of the heterogeneous cell surface glycans is a key requirement to address cell function and related cell biological processes. This project aims to develop a new platform technology for multiplexed glycan mapping of the surface of a single cell to address challenges of functional glycomics by utilising a conceptually new nanostrategy. By combining newly designed plasmonic nanoparticles with different surface-enhanced Raman scattering tags and multiple specific carbohydrate-recognising lectins, this project will produce a generic technology that is capable of non-destructive barcoding of the surface glycan signature of single cells in their native state and in response to metabolic perturbations.
Publication:
-Molecular Omics, 16(4), 339-344 (2020)
-Analyst, 149(6), 1774-1783 (2024)
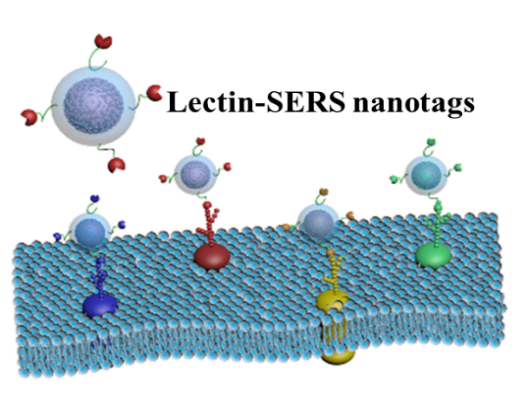
Collaboration with Dis Prof Nicki Packer (Macquarie University)
Biosensors
Due to the rarity and low amount of biomarkers for diagnostics, the development of sensor platform with the features of high sensitivity and multiplexed capability has attracted much attentions. In this project, we aim to develop a simple and rapid sensor platform for protein and gene biomarkers detection by combining the assay, e.g. microfluidic, lateral flow assay, magnetic beads, with the unique optical properties of nanomaterials.
Publications:
-Journal of Physical Chemistry B, 129(18), 4373-4382 (2025)
-Chemical Communications, 61(49), 8915–8918 (2025)
-ACS Applied Materials and Interfaces, 16(18), 22761-22775 (2024)
-ACS Sensors, 8(4), 1404-1421 (2023)
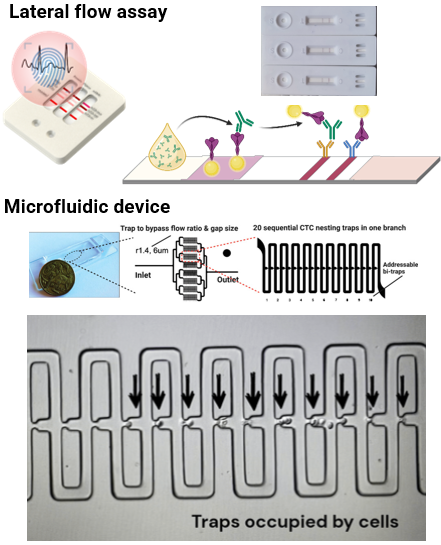
Collaboration with A/Prof David Inglis (Macquarie University)