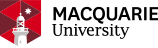Recombinant Protein and Antibody Characterisation
Recombinant Protein and Antibody Characterisation
Characterisation of antibodies/recombinant proteins
Case Study for Mass Spectrometry characterisation of a monoclonal antibody
APAF has expanded its protein analysis services for analytical characterisation of monoclonal antibodies including intact mass analysis, peptide mapping, and N-linked glycan profiling. This case study shows an example of these services.
Sample: Waters Intact mAb standard, Part # 186006552
The amino acid sequence of the antibody:
Heavy chain:
QVQLKESGPGLVAPSQSLSITCTVSGFSLLGYGVNWVRQPPGQGLEWLMGIWGDGSTDYNSALKSRISITKDNSKSQVFLKMNSLQTDD
TAKYYCTRAPYGKQYFAYWGQGTLVTVSAAKTTPPSVYPLAPGSAAQTDSMVTLGCLVKGYFPEPVTVTWNSGSLSSGVHTFPAVLQSDL
YTLSSSVTVPSSTWPSETVTCNVAHPASSTKVDKKIVPRDCGCKPCICTVPEVSSVFIFPPKPKDVLTITLTPKVTCVVVDISKDDPEVQFSW
FVDDVEVHTAHTQPREEQFNSTFRSVSELPIMHQDWLNGKEFKCRVNSAAFPAPIEKTISKTKGRPKAPQVYTIPPPKEQMAKDKVSLTCM
ITDFFPEDITVEWQWNGQPAENYKNTQPIMDTDGSYFVYSKLNVQ KSNWEAGNTFTCSVLHEGLHNHHTEKSLSHSPG
N-terminal modification Q > PyoGlu: -17 Da
Predicted Heavy chain average mass: 48484 Da
Light chain:
DVLMTQTPLSLPVSLGDQASISCRSSQYIVHSNGNTYLEWYLQKPGQSPKLLIYKVSNRFSGVPDRFSGSGSGTDFTLKISRVEAEDLGVYYC
FQGSHVPLTFGAGTKLEIKRADAAPTVSIFPPSSEQLTSGGASVVCFLNNFYPKDINVKWKIDGSERQNGVLNSWTDQDSKDSTYSMSSTLT
LTKDEYERHNSYTCEATHKTSTSPIVKSFNRNEC
Predicted Light chian average mass: 24198 Da
17 disulphide bonds, -34 Da
Intact protein average mass based on amino acid sequences: 145330 Da.
Intact mass analysis
Intact protein mass measurement is an important analytical characterisation technique that can be accomplished using mass spectrometry. The method is used to determine the homogeneity of recombinant or purified protein preparations, to confirm predicted molecular mass and identify deviations that could be due to cleavages and amino acid modifications. For the monoclonal antibody sample tested here, the intact mass was measured using an AB Sciex Q-ToF mass spectrometer coupled with an Agilent 1100 HPLC. The multiply charged MS spectrum of the antibody (Fig. 1) is de-convoluted to show the intact masses (Fig. 2) of the sample. The observed intact masses are the sum of the amino acid sequence mass of the protein (145330 Da) and the glycan masses (about 2900 Da to 3550 Da). This intact mass analysis result confirms that the antibody is pure and is heterogeneously glycosylated.
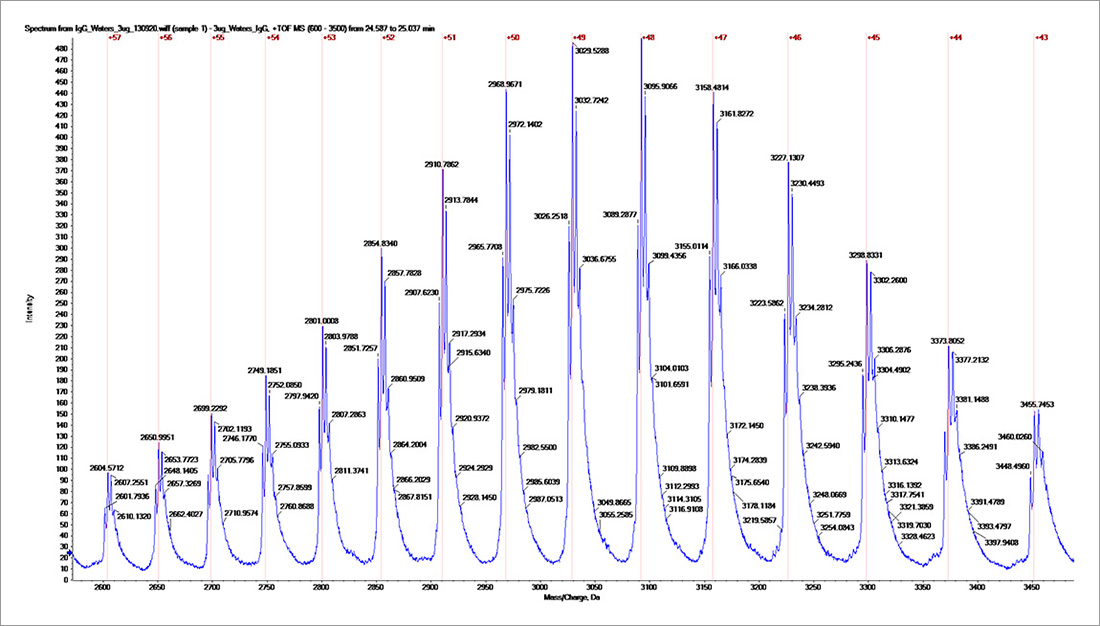
Fig. 1, Multiply charged intact mass spectrum for the IgG.
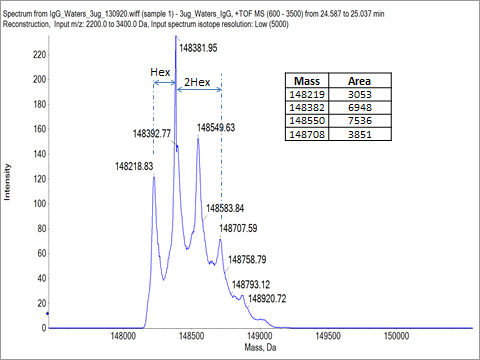
Fig. 2. Deconvoluted intact mass spectrum.
Glycan profile analysis:
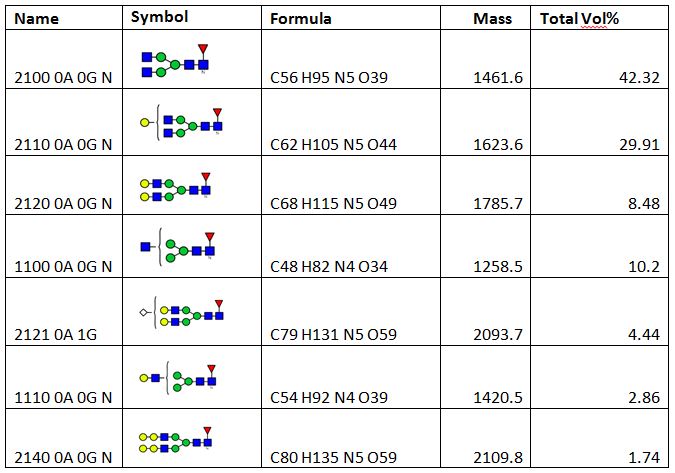
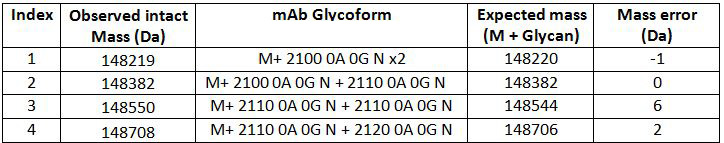
Glycan profile analysis was conducted using mAb-Glyco chip LC-MS system (Agilent Technologies). The result shows the relative abundances of observed N-linked glycans for the antibody. For the tested antibody, the main glycans detected are shown in Table 1. The result shows the top five glycan forms are of more than 95% abundance. As these top five glycan forms are of masses between 1258.5 Da to 2093.7 Da, the two heavy chains of the antibody must be both glycosylated to account for the intact masses of the protein being between 148235 Da to 148864 Da (AA mass of the protein 145330 Da plus two glycan masses). Table 2 shows the intact mass peak assignment based on the glycan profile analysis result.
Table 1, Main glycan forms and their relative abundances for the tested antibody
Table 2, Intact mass peak assignment based on glycan profiling result
Peptide mapping:
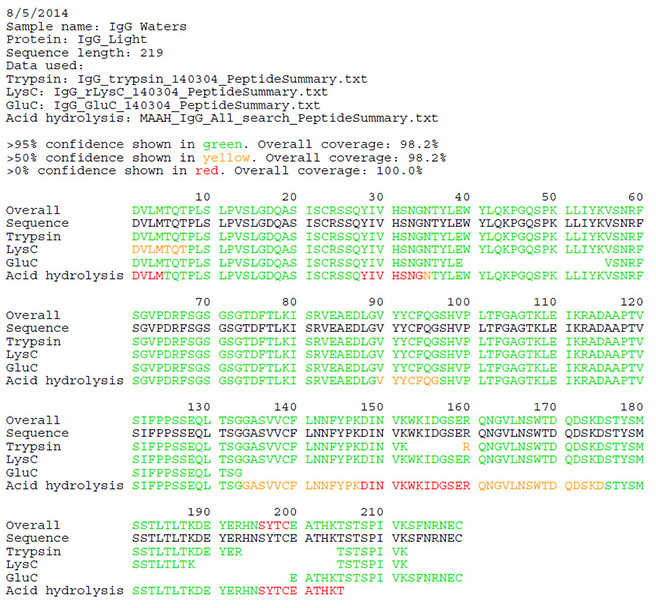
The antibody sample was digested using 3 different enzymes (Trypsin, Glu C, Lys C) and microwave assisted acid hydrolysis after reduction and alkylation. The peptide MS/MS data were acquired using high resolution, high mass accuracy mass spectrometer (Triple TOF 5600 or Orbitrap Elite). The MS/MS spectra were matched against the predicted peptide fragments for the antibody heavy chain and light chain amino acid sequences. The peptide mapping coverage was reported by combining peptides identified from all methods.
Light chain peptide mapping report:
Heavy chain peptide mapping report: