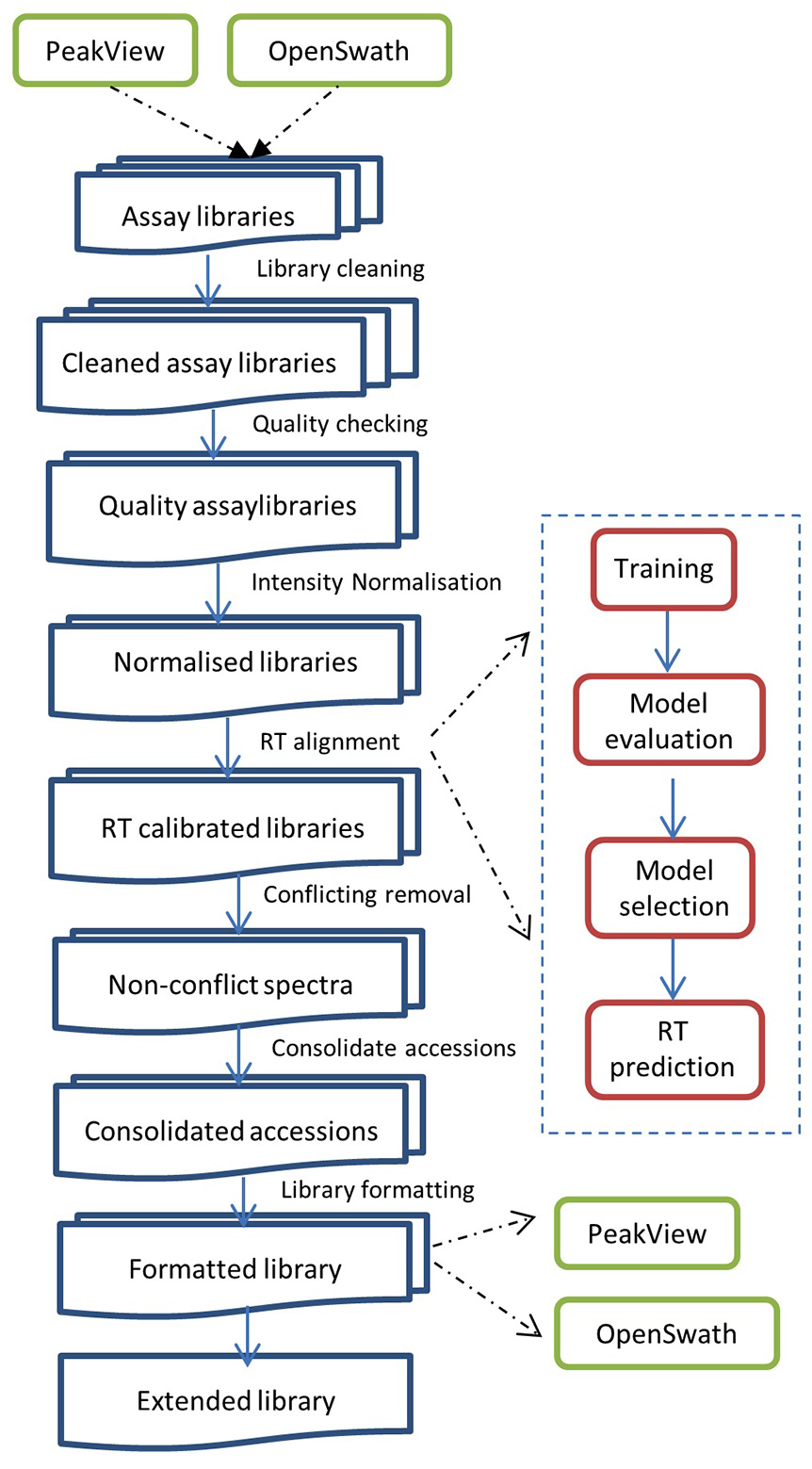
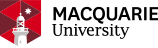SWATHXtend workflow
SWATHXtend workflow takes a seed library, which is usually a locally generated spectra library, and one or more add-on libraries which are subjected to an optional cleaning process to remove low confidence and low intensity peptide ion spectra. The cleaned libraries undergo a quality checking to ensure that the add-on library and the seed library have good matching quality in terms of retention time and relative ion intensity. All quality libraries undertake ion intensity normalisation and supervised learning based retention time alignment. For peptides of conflicting or overlapping spectra between seed and add-on libraries, only seed library spectra are kept. Protein accessions are consolidated by merging duplicated protein accessions in heterogeneous formats. The extended libraries can be output in PeakView or OpenSwath compatible format.