Quantitatitive and Comparative proteomic analysis using Mass Spectrometry
There are numerous techniques available now to identify protein differences in biological samples – known as comparative proteomics. With cell lines SILAC (Stable isotope labelling by amino acids in cell culture) can be used for metabolic labelling of proteins, although the “heavy” amino acids can be expensive. Chemical labelling techniques are an alternative approach. One of the earlier chemical labelling techniques for mass spectrometry analysis was ICAT (isotope-coded affinity tags) and this was quickly followed by the improved chemical labelling technique iTRAQ (isobaric tags for relative and absolute quantification). iTRAQ is the protein equivalent of transcriptomics.
iTRAQ labelling allows relative quantitation between 4 samples in each mass spectrometry run. To achieve this each sample is labelled at free-amines with specific iTRAQ reagents of a defined mass. A different iTRAQ reagent is used for each of the four samples. The samples are then mixed in equimolar amounts prior to mass spectrometry. For each peptide that is identified by mass spectrometry the iTRAQ label can be quantified. This provides the relative expression level of that protein across the four samples studied.
For cell lysate or tissue samples we conduct 2D-LC/MS which typically generates quantitative information for over 1500 different proteins. APAF has extensive experience with iTRAQ experiments, having used the technique since 2007. Many of our clients use iTRAQ as their first proteomics experiment to characterise their biological systems.
In a recent study with Dr. P. Scott Chandry of CSIRO Division of Animal, Food and Health Sciences, Werribee, Victoria, Australia we used iTRAQ quantitative proteomics and compared strains of two Salmonella enterica subspecies salamae serovar Sofia, one typically isolated from chickens and the other from a human. Salmonella Sofia is the most frequently isolated type of Salmonella in Australian chickens but interestingly it is not a pathogen. The Sofia serovar of Salmonella is only observed in Australia. This study was designed to identify proteins that may give clues as to how the Salmonella Sofia from chickens differs from those that occur in humans and to help our understanding of why this organism is so successful at colonizing chickens but is not pathogenic to humans. The research is currently being prepared for publication.
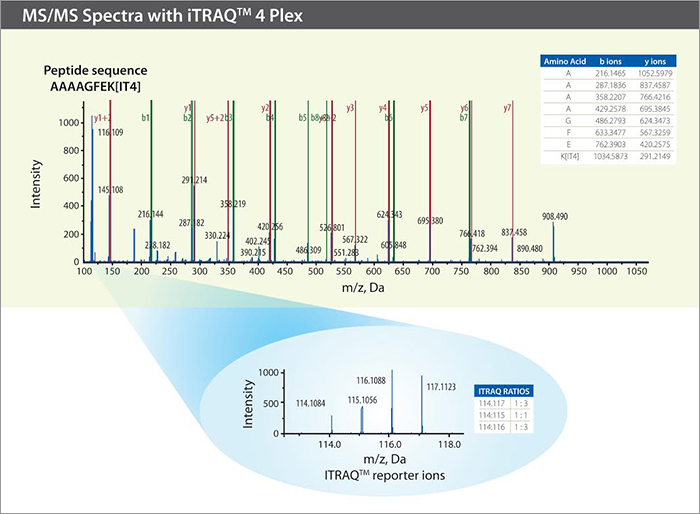
The graphic below is an example peptide MS/MS spectra from an iTRAQ experiment. The peptide b and y ions are used for identification, while the iTRAQ reporter ions at m/z 114-117 are used to determine the relative peptide quantities.
To get an indication of how the results are presented we have a results page available.