Protein Protein Interactions
Identify novel protein-protein interactions with “Pull Downs” or Co-Immunoprecipitation and Mass Spectrometry
Co-Immunoprecipitation, (Co IP) is a technique used to isolate protein complexes through the use of affinity interactions. Co-IP is typically used in a serial manner to evaluate interactions one protein at a time. It requires advanced knowledge of the likely interacting partners and a useful antibody to blot for its detection. As an alternate, mass spectrometry can be used to determine the identity of co-immunoprecipitated proteins. The clear advantage is that no prior knowledge of the likely interacting partners are required. By targeting a known member with an antibody and utilising mass spectrometry it may become possible to pull the entire protein complex out of solution and thereby identify unknown members of the complex. This concept of pulling protein complexes out of solution is sometimes referred to as a "pull-down". Co-IP is a powerful technique that is used regularly by cell biologists to analyse protein–protein interactions. The bait protein may be bound by an antibody, or be a protein construct that can be affinity isolated through a moiety or domain.
A Co-Immunoprecipitation experiment performed at APAF was carried out to identify novel S100A4 protein interactions in SW480 colon cancer cells. The workflow involves conducting a co-IP enrichment, washing the bound complex, eluting the complex, separating protein constituents by SDS-PAGE, in-gel tryptic digestion, and finally, LC/MS/MS. An iso-type control pull-down is used to ensure specificity of interactions.
Method
Cells were lysed in modified RIPA buffer (RIPA buffer containing no EGTA/EDTA). Two conditions were set up in the cell lysate; 1 mM EGTA or 2 mM CaCl2. Dynabeads (Protein G) (Invitrogen) were conditioned in NT-80 buffer (20 mM Tris-HCl, 80 mM NaCl) and used for preclearing. Conditioning consisted of incubating the beads in NT-80 buffer at RT for 5 min (three times). Preclearing consisted of incubation of the beads with cell lysate for 15 min at RT. Supernatant was retained and incubated for 2 h at 4 ˚C with 10 µg of either S100A4 (mouse, monoclonal) antibody (catalog #120-93283) (Abcam), or an IgG murine control (Sigma). Dynabeads (10 mg) were added and incubated for 2 h at 4 ˚C. Beads were washed twice in solution C (50 mM Tris-HCl, 0.2 % Triton X-100, 150 mM NaCl) followed by three washes in NT-80 buffer. 100 µl ammonium hydroxide (pH 11.5) was added to the beads and supernatant collected. Samples were dried by vacuum centrifugation (SpeedVac SC110A, Thermo) and stored at -20 ˚C until use for SDS-PAGE.
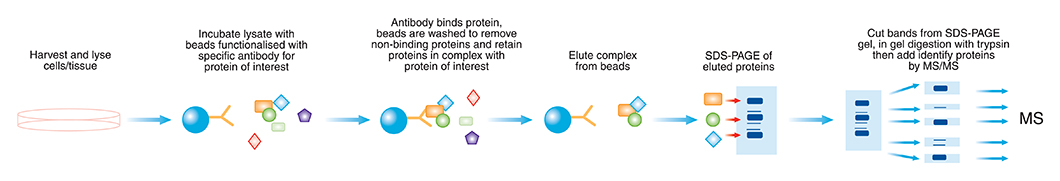
Results
Precipitate from the calcium condition co-immunoprecipitation with S100A4 antibody and IgG control was run on a gel and protein bands of interest excised. The Mascot search engine (Matrix Sciences) was used to identify peptides. Some of the proteins detected are shown in Table 12, which lists only those proteins unique to the S100A4 antibody co-immunoprecipitation (i.e. Proteins in Table 12 were not found in the IgG control).
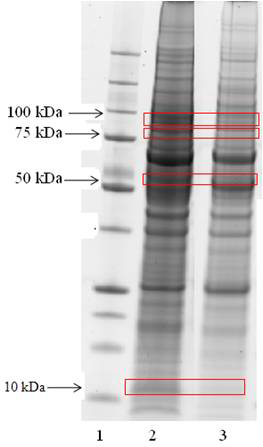
Precipitate from the S100A4 antibody co-immunoprecipitation (lane 2) and precipitate from the IgG control co-immunoprecipitation (lane 3). Bands of interest are indicated by red boxes. Candidates identified that are involved in cytoskeleton or cell cycle are mentioned below.
T-complex protein (TCP) essential for the correct folding of proteins, Annexin II has a biological role in cell division and apoptosis, Fascin an actin-bundling protein suggested to be involved in the assembly of actin filament bundle